Open Anatomy technical details
The Open Anatomy Project technology stack is being designed from the ground up using open technologies to promote colloborative development by distributed teams of researchers. This section provides some details about the different tools that make up Open Anatomy, our plans for the future, and how we see technology choices influencing the kind of collaborative community we want to build.
The Open Anatomy paper provides additional detail into the design of the current data format and atlas viewer: The Open Anatomy Browser: A Collaborative Web-Based Viewer for Interoperable Anatomy Atlases, Halle M, Demeusy V, Kikinis R. Front Neuroinform. 2017 Mar 27;11:22. doi:10.3389/fninf.2017.00022.
Architecture
Open Anatomy consists of several basic building blocks that work together to allow users to create, view, and share anatomy atlases:
- an atlas data format for representing atlases as annotated collections of images and geometry, along with visual styles for display;
- atlas viewers and other software to access atlases;
- authoring tools for creating, editing, and extending atlases;
- a collaborative web-based portal that serves as a central location to find existing atlases and develop and publish new ones.
Similar types of collaborative tools, built on the same kinds of components, have had transformative impact in other fields. Wikipedia has transformed encyclopedias into communal sources of knowledge. In the software development world, GitHub has been pivotal in making code sharing and distributed development of complex software possible. Wikipedia deals with text articles and pictures, while GitHub deals with files and computer code. Each have specialized tools to deal with their specific kinds of data, and each uses web-based standards and web browsers to access that data.
In much the same way, Open Anatomy deals with models, imaging data, and textual annotations, and visual styles to display them. Let’s look more in detail at the different components of the stack.
Atlas data format
Typically, the structures that make up an atlas are described by their shape (geometry represented as 3D models or voxels), their semantic meaning (text labels or more complex ontological annotations), their appearance (styles analogous to CSS), and their relationship to other data (underlying imaging from CT, MRI, or microscopy, or perhaps spatially organized quantitative measurements).
No common open data format exists to describe the information that makes up atlases. We are closely collaborating with members of the anatomy and neuroinformatics communities (such as the Human Atlas Working Group) to develop a data format that can use used for atlas interoperability and exchange.
Based on these discussions, we have implemented a draft file format based on JSON-LD for representing atlases and tying together imaging and geometry data represented in existing file formats, much like HTML references media files using the img or video tags.
This atlas data format remains under active development. We believe several design features are important, however:
- contributors with different skills (anatomy, medicine, labeling, illustration) should be able to work together effectively,
- the contribution of every contributor should be specifically acknowledged,
- atlases should be modular and extensible in a way that allows reuse without wholesale copying or the permission of the original creators (this property gives GitHub forks their power),
- atlases should provide web-standard linking mechanisms to associate anatomic structures with external information.
One of the most important areas of modularity we wish to address is adding new annotations to an existing atlas, specifically translations into new human languages. We also want to use annotations to provide links to existing repositories of online medical knowledge, including Wikipedia.
Status: draft format
Why another format?
The atlas data format we are developing fills a gap that no other format addresses. Our format allows us to add web- and RDF-like semantic information spatial descriptions stored in image data and geometry files. Identifying features in images and describing their shape is a primary task in imaging sciences including medical imaging. Each field has their own particular data formats to represent those features.
What those formats don’t generally provide is a way to uniquely refer to those features from outside the file. For instance, label maps are common in medical imaging, where the shape of structures is described by a per-voxel integer label, with each integer corresponding to one structure. This image-based format can describe detailed anatomical structures, but offers no way to, say, attach a relationship described by an RDF triple to a single structure. The same problem occurs in text processing: there is no HTML-level way of referring to content in an HTML file without anchors being defined.
Our data file effectively provides anchors that allow semantic information to be associated with structures defined inside images and geometry. It associates data accessor objects that uniquely identify image and geometry features in external data files with concept nodes. These concepts can be referenced by URI, and thus can be linked to using URLs and described using RDF-style metadata.
In fact, we believe that the underlying data format should be as generic as possible, without requiring anatomy- or medical semantics. This abstraction opens the possibility of using the same format in other imaging fields, such as microscopy, astronomy, and satellite imaging.
Our overriding philosophy is to avoid duplicating the information contained in any existing file format, but instead associating the information contained within them in new and different ways.
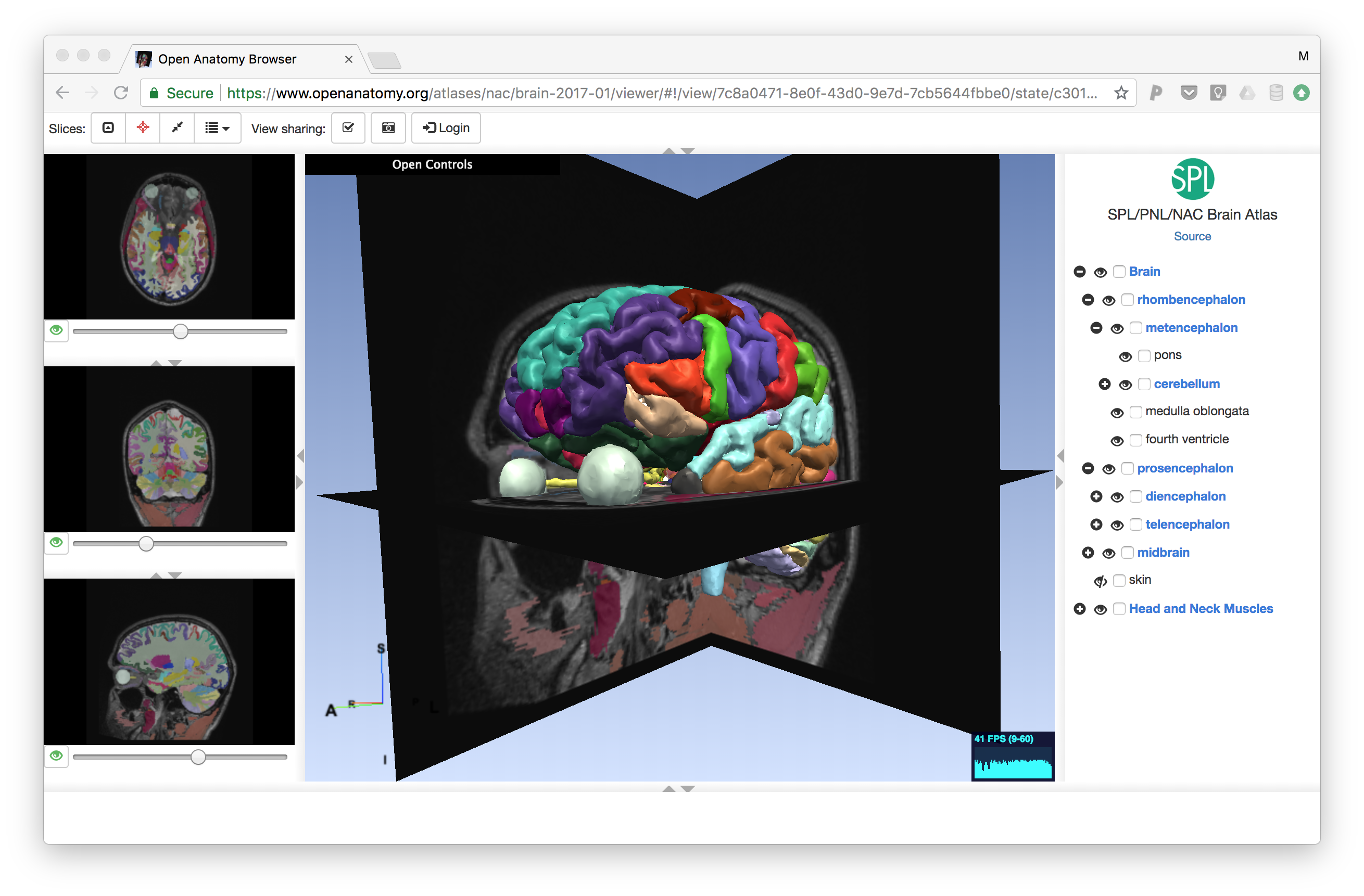
Atlas viewers
Atlases are naturally visual, and require visual tools to view. To validate our atlas data format, we have developed a prototype quality anatomy atlas viewer called the Open Anatomy Browser (OABrowser). OABrowser uses WebGL through the THREEJS library and is currently built using Angular 1 application architecture and UI components.
Currently, OABrowser is designed for desktop use and hasn’t been optimized for mobile viewers. We are planning the design of the next generation viewer that will include mobile support, at least at the tablet level.
We intend for there to be many atlas clients. A common atlas data format means that anyone can write an atlas viewer, perhaps specializing it for different kinds of data. For example, a microscopy-based atlas would be designed to view the very large images common in the field. Atlas client software can be non-interactive as well: for instance, automated testing clients could scan atlases looking for inconsistent data, or data mining software could use atlases as a spatial index into larger medical data sets.
Specialized atlas viewers could be designed for classroom teaching. The OABrowser has a simple scene sharing mechanism: paste the URL from the viewer into another browser, and the two will stay synchronized. While this feature is a proof of concept, we anticipate the development of future collaborative atlas viewers. View sharing is currently implemented using Google’s Firebase real-time database.
Status: prototype implementation
Authoring tools
We need tools to create great atlases. Initially, we use 3D Slicer to author atlases using manual post-processing of Slicer MRML files. We are currently implementing atlas export from 3D Slicer. Finalizing an atlas data format will allow other software tool builders to make new atlases, and remains a high priority.
But authoring can mean more than creating new atlases from scratch. Even simple acts such as sharing a bookmarked view, changing a color scheme, or adding new labels or translations is authoring. Our goal is to pursue web-based editors for these simpler authoring tasks while we simultaneously look towards atlas export from major medical software programs.
Status: in development.
Backend portal
Atlases don’t require a portal or dedicated server software. They can be viewed as files on disk or served from any standard web server. (In fact, the current implementation does just that.)
But our larger goals of building a collaborative repository of atlas-based medical knowledge is furthered by a central focus and helpful infrastructure. GitHub is an good parallel. GitHub relies on the git distributed version control system as its data model. GitHub itself isn’t strictly necessary for distributed development. But GitHub provides an unparalleled social focus for software developers that has helped build hundreds of communities around software libraries.
An atlas data portal can also address data needs specific to anatomy atlases. Atlases are made up of distinct types of data with different characteristics and usage patterns. Annotation and text labels are relatively small and simple to deal with: they act like text, and we have lots of experience storing, differencing, and versioning text. Geometric information, image data, and per-voxel labels are larger and are very different from text, thus are more difficult to handle.
Luckily, geometry and image data aren’t edited as much as text data. Also, if multiple atlases share geometric models, browsers can cache that data and avoid loading it more than once.
An additional feature of a backend portal provided by both Wikipedia (through Talk pages) and GitHub (through Issues) is the ability to discuss issues related to specific artifacts such as articles or code. Anatomy includes discussion and debate. We expect lively discussion to improve the quality of atlases and lead to new ideas and atlas development.
Finally, backends can enforce community rules and standards. Anatomy atlases are used for medical purposes. It is not unreasonable to expect that community standards and protections may emerge to prevent incorrect or malicious information from being disseminated. GitHub’s model of authentication and permissions mitigate this problem somewhat. We anticipate the need for similar mechanisms on top of which approval and review tools could be build.
Status: in design. The Open Anatomy Project currently does not include a dedicated backend server.
Inspirations
The design choices of Open Anatomy is informed heavily by other open source and community-driven websites and technologies.
3D Slicer
We are strongly influenced by the design and development methodology used to create and maintain 3D Slicer, our open-source medical image computing and visualization platform. 3D Slicer was created as in in-house project at the Surgical Planning Lab at Brigham and Women’s Hospital to allow medical researchers to share their work, and to enable visiting scientists and physicians to continue collaboration once they left the SPL. Slicer 4, the latest version of the software, is developed and tested using industrial-strength methodologies developed with assistance from Kitware Inc.
Slicer4 is maintained by a group of researchers and engineers throughout the world, paid for primarily through a collection of grants from the National Insitututes of Health (NIH) and other sources. It is licensed under liberal terms (the 3D Slicer License) that allow for any use, including commercialization, without cost.
Slicer4 uses a file format called MRML to store information about medical images, geometry, labels, and geometry. We have learned many lessons from MRML and have used them to guide design decisions for the atlas data format.
Wikipedia and GitHub
We have already discussed technical similarities between Open Anatomy, Wikipedia and GitHub. But these sites and the communities behind them serve a more fundamental purpose: an existence proof for the human desire to pool information. When Wikipedia first came on line, it was an open question whether volunteers would contribute their time to create high quality encyclopedic articles about, well, everything. Now, millions of articles in hundreds of languages later, then answer is yes. GitHub as well: vibrant communities have sprung up around tens of thousands of software projects, big and small. Some are maintained by a single volunteer.
Call to action
The willingness to give freely for the good of others is part of the hypothesis of the Open Anatomy Project. Other projects have shown the way. Hard work lies ahead.
We are just beginning to build this larger vision. There are lots of moving parts, both technical and social. We need help from you to make the technical and educational vision of Open Anatomy a reality.