History
The Open Anatomy Project has come out of years of medical image computing research at the Surgical Planning Laboratory (SPL) in the Department of Radiology at Brigham and Women’s Hospital, a teaching affiliate of Harvard Medical School. The SPL, founded by Dr. Ron Kikinis in the early 1990’s, has been a pioneer interdisciplinary laboratory that has combined the strengths of clinical medicine, radiology, and computer science. Physicians and computer scientists from around the world have come to the SPL to analyze and visualize their medical data and apply the resulting knowledge to help treat patients.
The SPL and open development
At the SPL, we have learned a great deal about the power of collaboration and open sharing of ideas and software. We have developed 3D Slicer, a cross-platform open source software tool for medical image analysis and visualization. 3D Slicer version 4 has been downloaded over 400,000 times since its release in 2011 and has one of the largest developer and user communities of any open source medical software tool.
Over the years, researchers at the SPL have used 3D Slicer and similar tools to segment and annotate complex medical data sets derived from medical imaging as part of their research. These data sets were originally designed for applications such as surgical planning, atlas-based registration of patient data, the understanding of diseases such as schizoprenia, and other purposes. The development of these data sets at the SPL, many of which we now distribute as atlases, would not have been possible without the consistent data management of Marianna Jakab.
Atlas viewers
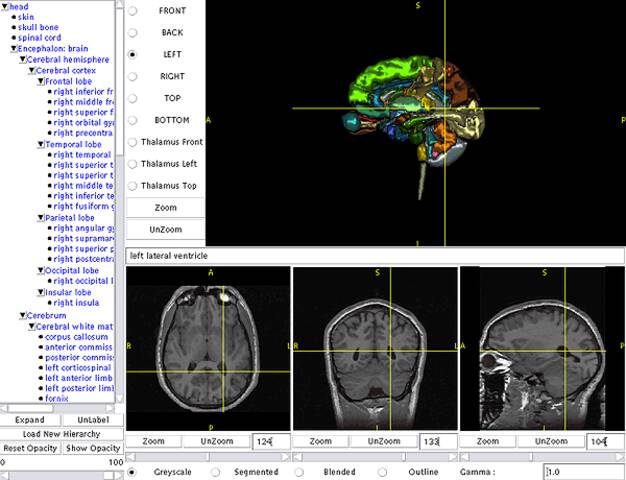
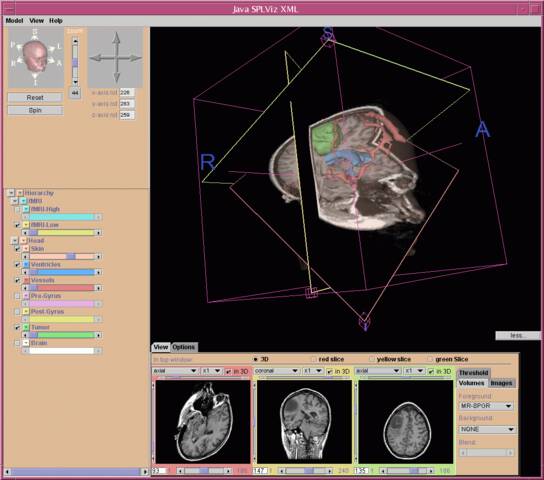
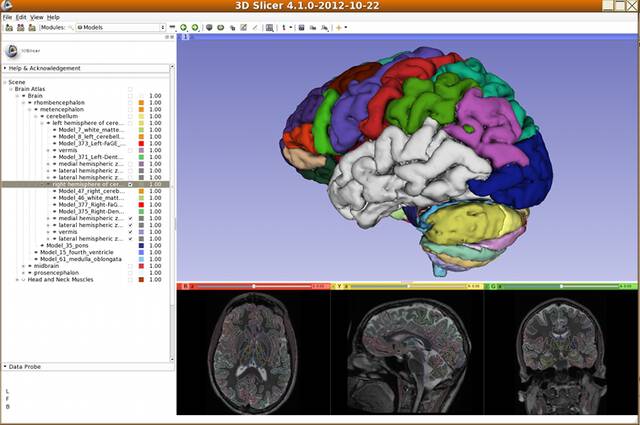
Dr. Kikinis realized early on that atlases based on these detailed data sets would be valuable for medical education. Working with medical colleagues such as Dr. Martha Shenton and computer scientists such as Chris Umans, Dr. Michael Halle and Dr. Polina Golland, they developed one of the first web browser-based anatomy atlases in 199812. The SPL developed a Java-based 3D atlas viewer called SPLViz in 2001.
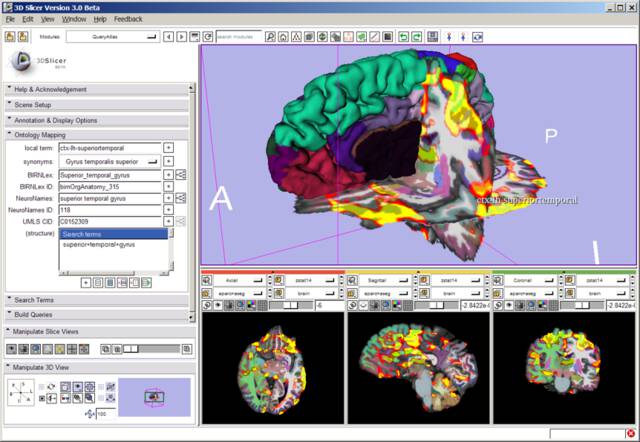
Other atlases developed at the SPL required 3D Slicer to view, which offered great power to the user but limited the audience who could access the atlas. The SPL’s Query Atlas for Slicer 3 featured three-dimensional views integrated with structure labels from various medical ontologies.
As part of the Neuroimage Analysis Center funded by the National Institute of Biomedicine and Bioengineering (NIBIB), Dr. Halle and his team has worked to curate the SPL’s brain atlas. Developed as a tool for advanced segmentation and registration algorithms, the SPL/NAC Brain Atlas consists of hundreds of individual brain structures. NAC’s Atlas Infrastructure Core developed new tools for organizing and viewing the brain atlas.
In 2016, the Open Anatomy Project was started based on the tools, data, and insights that came from the NAC Atlas Infrastructure Project. Open Anatomy goes beyond NAC’s neuroscience research mandate to build atlases of other parts of the body. The Open Anatomy Project includes new collaborative atlas development, outreach, and educational components.
Beyond the body: astronomy
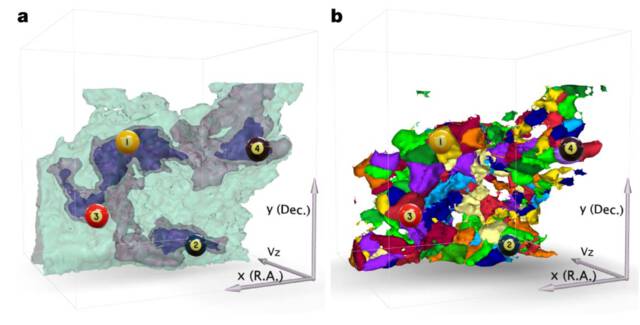
Members of the Open Anatomy team have used the same tools and techniques used for human anatomy to identify meaning in other kinds of scientific image data. Beginning in 2005, Dr. Halle and Dr. Kikinis worked in collaboration with a team from the Harvard-Smithsonian Center for Astrophysics (CfA) to use medical segmentation and visualization algorithms to understand the structure of the IC 348 star-forming complex in Perseus. The Harvard group included Professor Alyssa Goodman, researchers Michelle Borkin and Jens Kauffmann, and other colleagues.
Using 3D Slicer and other tools, the team was able to visualize the three-dimensional structure and organization of the star-forming regions, identify gas jets and outflows,and better understand the role of gravitation in star formation3. This collaborative work in cross-disciplinary research became a core project of Harvard University’s Initiative in Innovative Computing (IIC).
The “Astronomical Medicine” project bridged two very different areas of science by exploiting commonality between medical imaging and astronomy. Both fields are heavily dependent in multi-dimensional imaging. Both use extra dimensions of imaging (multiple wavelengths or modalities) to detect relevant features. And both fields identify and extract those features from the raw imaging data, allowing them to be further analyzed and visualized.
In 2009, the group published their results, “A role for self-gravity at multiple length scales in the process of star formation”4 in the journal Nature. As an electronic supplement to the journal, a 3D PDF file provided an interactive view of the figures in the paper. This figure was labeled with markers that identified the relevant structures.
These lessons have helped shape the approach taken by the Open Anatomy Project. Open Anatomy was designed with applications beyond anatomy atlases, like astronomy, in mind. Based on our interdisciplinary work with the CfA and other groups, we have created a data format that can used in the broad range of imaging applications that identify locations or regions of interest in imaging data or geometry. Our format doesn’t specify a particular file format for storing images, so it can be compatible with medical standards such as DICOM or NIfTI as well as astronomy’s FITS files. It is also compatible with different metadata vocabularies for labeling features or regions used in different fields.
The Surgical Planning Lab’s interdisciplinary history has provided the Open Anatomy Project an excellent foundation for future applications outside of anatomy atlases. Our goal is to enable researchers to publish their own structured image data like the Astronomical Medicine’s figure from Nature, along with the underlying data, in a web browser for anyone to explore. We hope to apply our tools to other imaging areas including microscopy and remote sensing.
References
-
Golland P., Kikinis R., Umans C., Halle M., Shenton M.E., Richolt J.A. (1998) AnatomyBrowser: A framework for integration of medical information. In: Wells W.M., Colchester A., Delp S. (eds) Medical Image Computing and Computer-Assisted Intervention — MICCAI’98. MICCAI 1998. Lecture Notes in Computer Science, vol 1496. Springer, Berlin, Heidelberg ↩
-
Anderson J.E., Umans C., Halle M., Golland P., Jakab M., McCarley R.W., Jolesz F.A., Shenton M.E., Kikinis R. Anatomy Browser: Java-based Interactive Teaching Tool for Learning Human Neuroanatomy. Radiological Society of North America - Electronic Journal 1998. ↩
-
Demonstration of the applicability of 3D Slicer to Astronomical Data Using 13CO and C18O Observations of IC348. Michelle A. Borkin, Naomi A. Ridge, Alyssa A. Goodman, Michael Halle. arXiv, 24 Jun 2005. ↩
-
A role for self-gravity at multiple length scales in the process of star formation. Alyssa A. Goodman, Erik W. Rosolowsky, Michelle A. Borkin, Jonathan B. Foster, Michael Halle, Jens Kauffmann & Jaime E. Pineda. Nature volume 457, pages 63–66 (01 January 2009). ↩